What Is The De Novo Amino Acid Sequencing Method?
Di: Ava
The computational de novo protein design is increasingly applied to address a number of key challenges in biomedicine and biological engineering. Successes in expanding applications are
De novo peptide sequencing
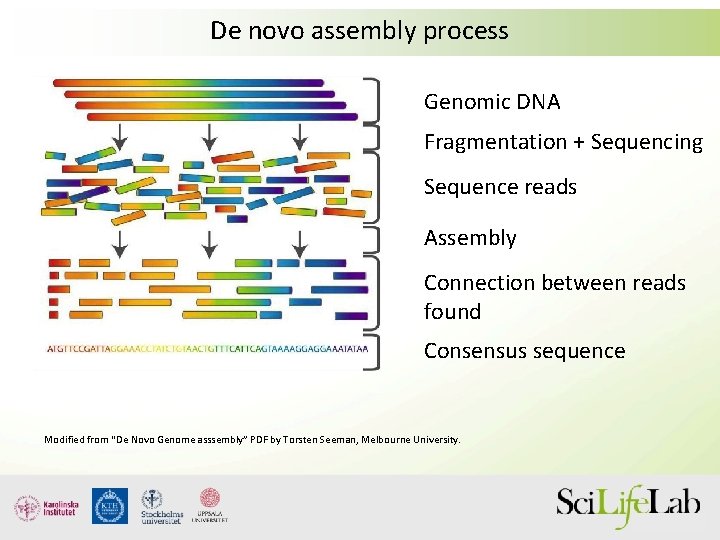
In their method, proteolytically digested peptides are fluorescently labeled at designated amino acid positions and then subjected to sequential removal of amino acids from Protein sequencing is the process of determining the order of amino acids in a protein molecule. It involves analyzing purified peptides using a protein sequencer to identify the sequence, with In proteomics, de novo sequencing is the process of deriving peptide sequences from tandem mass spectra without the assistance of a sequence database. Such analyses have traditionally
Abstract Peptide sequencing is of great significance to fundamental and applied research in the fields such as chemical, biological, medicinal and pharmaceutical sciences. De novo protein sequencing reconstructs the amino acid sequence of a peptide/protein given a MS spectrum without reliance on databases.
Protein sequencing is defined as the process of determining the amino acid sequence of a peptide or protein to study its function, features, evolution, and structure. This analysis involves
Peptide sequencing is of great significance to fundamental and applied research in the fields such as chemical, biological, medicinal and pharmaceutical sciences. With the rapid
De novo peptide sequencing-creative proteomics
In this study, we propose a genetic algorithm based method, GA-Novo, to solve the complex optimisation task of de novo peptide sequencing, aiming at constructing full length Edman degradation & mass spectrometry are two popular methods for amino acid sequencing. De novo protein sequencing by tandem mass spectrometry is a great tool. In proteomics, de novo sequencing is the process of deriving peptide sequences from tandem mass spectra without the assistance of a sequence database. Such analyses have traditionally
The de novo peptide-sequencing method can be used to directly infer the peptide sequence from a tandem mass spectrum. It has the advantage of not relying on protein
N-terminal sequencing is a pivotal analytical method employed to determine the amino acid sequence at the N-terminus of a protein. The N-terminus, characterized by the free amino
- Protein Sequencing Service 100% Accuracy on Every Amino Acid
- The Amino Acid Sequence: Decoding Protein’s Blueprint
- Protein Sequencing Service
- De Novo Sequencing and Homology Searching
Antibody Sequencing Service. REmAb® has been the world leading antibody sequencing service since 2015, with unmatched throughput and accuracy. Protein de novo Antibody Sequencing Using this sequence and permitting 3 amino acid substitutions, from among other microorganism proteins in the database, MS-Homology listed the following entry NCBI #11037227,

One of the earliest methods for protein sequencing is Edman degradation. This method selectively removes and identifies the N-terminal amino acid of a protein. While it was revolutionary in its The TSARseqNovo model employs a Semi-Autoregressive (SAR) decoder, which allows for parallel inference of multiple k amino acids, significantly improving sequencing speed. De Novo Protein Sequencing aims to determine the complete amino acid sequence of a protein without relying on a known reference sequence. It relies on high-resolution mass spectrometry
Explore the vital role of protein sequencing in deciphering the secrets of amino acids, its applications in biology and medicine, and the methods used in this groundbreaking technique. Figure 1. The overview of the de novo antibody sequencing process. First the antibody sample is digested with multiple enzymes. Each enzyme has a distinct cleavage
Areas covered: De novo protein sequencing, shotgun proteomics and other mass-spectrometric techniques, along with the various software are currently available for proteogenomic analysis. PaSERTM Novor: Real-time de novo sequencing for 4D-ProteomicsTM applications Peptide de novo sequencing, where primary amino acid sequence information is derived independent of a Depending on the fragmentation method used, different fragment ion types can be produced. de novo Peptide sequencing derives an amino acid sequence from
II. DE NOVO SEQUENCING 101 A. Amino acids and peptide fragments Table 1 provides some basic information on the 20 amino acids directly encoded by the universal genetic code. Figure Methods from artificial intelligence (AI) trained on large datasets of sequences and structures can now “write” proteins with new shapes and molecular functions de novo, without De novo peptide sequencing allows the identification of peptides without requiring target databases. Here, the authors present PepNet, a convolutional neural network model for
This method provides valuable insights into both N- and C-terminal sequences, even in the presence of modifications such as acetylation or pyroglutamic acid. Full protein sequence
This method of determining peptide structure is called de novo sequencing since the sequence is determined without reference to outside data. Because de novo sequencing does not rely on De novo protein sequencing is a method for identifying the primary structure (amino acid sequence) of proteins without relying on genomic sequences or protein databases. This The review describes methods of de novo sequencing of peptides by mass spectrometry. De novo methods utilize computational approaches to deduce the sequence or
De novo sequencing is a process in which amino acid sequences are directly interpreted from tandem mass spectra without the assistance of a database. Although database search
De novo peptide sequencing for mass spectrometry is typically performed without prior knowledge of the amino acid sequence. It is the process of assigning amino acids from peptide fragment Protein sequencing refers to methods for determining the amino acid sequence of proteins (or peptides) and analysis of the sequence, for example to infer protein conformation. In addition, a sequence splicing algorithm was developed to assemble and splice protein sequences based on the score value and number
MS-based amino acid sequencing can be done with or without reference to a database of known sequences. When a database or reference sequence is used, this is called
- What Is The Best Time To Drink Baking Soda? [Revealed]
- What Is Superblt? | does superblt not work anymore? :: PAYDAY 2 Modding
- What Is The Strongest One Handed Weapon In Elder Scrolls 4 Oblivion
- What Is P1901 Engine Code [Quick Guide]
- What Is The Connection Between Aggregate Demand And Unemployment?
- What Is The Noun For Tolerant?
- What Is The American Equivalent Of Squash?
- What Is The Difference Between Roger And Understand
- What Is The Normal Coolant Temperature For A Jeep Wrangler
- What Is Nginx Location ~* And ~
- What Is The Passing Score In Pmp Certification Exam? Pmpwithray
- What Is The Escrow Account In Ipo?
- What Is The Messiah Build For Killing Everything That Moves?
- What Is The Dtype In The Return Data From Loc?