Single-Cell Rna-Sequencing In Neuroscience
Di: Ava
The ability to deeply phenotype different iPSC-derived cell types is therefore of primary importance to the successful and informative application of this technology. Here we describe a combination of motor neuron (MN) derivation and single-cell RNA sequencing approaches to generate and characterize specific MN subtypes obtained from A systematic collection for single-cell and spatial transcriptomics is critical for in-depth analysis and novel discovery in AD. Here, authors show ssREAD which covers over 7 million cells and 381
Single-cell RNA-Sequencing in Neuroscience
INTRODUCTION TO SINGLE CELL RNA-SEQ Analysis of single cell RNA-seq data – online course Katarzyna Kania (CRUK CI Genomics Core Facility) 04th November 2021 Together we will beat cancer Single-cell RNA sequencing (scRNA-seq) provides an alternative method to study the cellular heterogeneity of the brain 15, 16, 17, by profiling tens of thousands of individual cells 15, 18, 19. Single-cell RNA-sequencing (scRNA-seq) has added substantially to our knowledge about cellular diversity and signaling networks, particularly during stages of peak neurogenesis.
Hao et al. performed single-cell RNA sequencing with immunostaining and neurosphere cultures on adult macaques and revealed robust neurogenesis in the adult macaque hippocampus.
PEX5L, found to be highly oligodendrocyte specific in single-cell and single-nucleus RNA-seq 45, 50, was downregulated at both RNA and protein level. Single-cell suspensions from the hemorrhaged region of the ipsilateral striatum on day three post-ICH were profiled using single-cell RNA sequencing (scRNA-seq). Gene Ontology (GO) and gene set variation analysis (GSVA) further
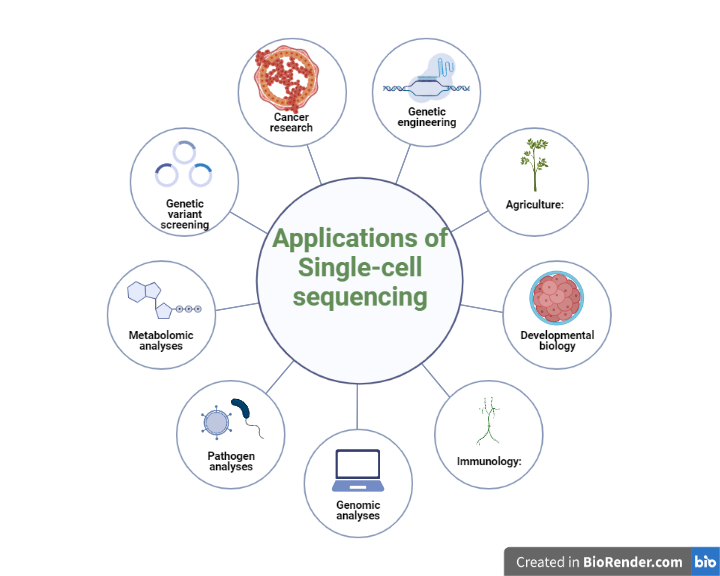
Single-cell sequencing, while a relatively recent development, is opening doors in our understanding of cell populations and tissues. This article explores how this technology works and what it tells us.
- Multimodal Nature of the Single-cell Primate Brain Atlas
- Parkinson’s disease in the single-cell era
- Single cell profiling and analysis in neuroscience
- Single-Cell RNA Sequencing Characterizes the Molecular
Single-cell RNA sequencing and spatial transcriptomics reveal a novel mechanism of oligodendrocyte–neuron interaction in cognitive decline after high-altitude cerebral edema.
We employed a droplet-based single-cell RNA-sequencing approach to develop a comprehensive census of molecularly distinct cell types Neuroscience research greatly benefits from single-cell sequencing technologies, which can reveal transcriptional alterations on a cellular level. However, preparing single-cell suspensions is technically challenging, requires experience, and has The central nervous system (CNS) comprises a diverse range of brain cell types with distinct functions and gene expression profiles. Although
Unraveling the complex pathophysiology of white matter
Single-cell analysis of mouse DRG This page contains additional data supporting our manuscript Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing (Published in Nature Neuroscience) Dmitry Usoskin [1], Alessandro Furlan [1], Saiful Islam [1], Hind Abdo [1], Peter Lönnerberg [1], Daohua Lou [1], Jens Hjerling-Leffler [1], Jesper
Single-cell RNA-sequencing (scRNA-seq) has since then opened the window to a deeper understanding of cellular identity and is becoming a widely used method in molecular biology. Integrative genetic and single cell RNA sequencing analysis provides new clues to the amyotrophic lateral sclerosis neurodegeneration Hankui Liu 1,2† Liping Guan 1,3† Min Deng 4† Lars Bolund 5,6 Karsten Kristiansen 3 Jianguo Single-cell sequencing approaches have revealed that gene expression patterns in the brain are cell-type specific, not only differentiating major classes of cells such as neuronal and glial cells

Single cell analysis is, in many ways, the harbinger of a broader transition towards “contextualized genomics,” in which the information-rich readouts of DNA sequencing can be leveraged to make many new kinds of measurements of cells and tissues. I will describe the ways in which this technological frontier is likely to impact neurobiology. The authors used single-cell genomics to profile thousands of human dopamine neurons and identify one uniquely Parkinson’s disease-susceptible population, which was enriched for genetic risk for
In this talk, I will introduce the basic experimental and computational concepts in single cell analysis, and provide some practical guidance on its application to problems in neuroscience. Here we contribute to this growing effort by applying single-cell RNA Sequencing (scRNA-seq) to characterize the transcriptomic profiles of tectal cells labeled by the transgenic enhancer trap line y304Et (cfos: Gal4;UAS: Kaede). We sequenced 13,320 cells, a 4X cellular coverage, and identified 25 putative OT cell populations.
Single cell RNA-sequencing technologies (scRNAseq) are rapidly developing and allow for the interrogation of activity-dependent transcription at cellular resolution. Single-cell sequencing technologies measure RNA or DNA from individual cells without the need for selective cell purification. These techniques can be summarized by three characteristics: scope (number of cells), granularity (number of genes or epigenetic features), and spatial resolution (Fig. 1b).
Single-cell RNA-seq and CITE-seq were used to profile the glioblastoma immune landscape in humans and mice, revealing the diversity and dynamics of tumor macrophages as the disease progresses from
Emerging technologies such as single-cell RNA sequencing (scRNA-seq) and Spatial Transcriptomics (ST) are potent tools for exploring the molecular complexity, cell heterogeneity, and functional specificity of CNS disease. scRNA-seq and ST can provide insights into the disease at cellular and spatial transcription levels.
Abstract Single-cell sequencing technologies, including transcriptomic and epigenomic assays, are transforming our understanding of the cellular building blocks of neural circuits. By directly measuring multiple molecular signatures in thousands to millions of individual cells, single-cell sequencing methods can comprehensively characterize the diversity of brain
Single-cell RNA sequencing has advanced our understanding of cell-specific expression in complex tissues, such as brain.101112131415 To isolate individual cells, the tissue is dissociated, resulting in destruction of the tissue architecture and gene expression artifacts. 16 Spatial transcriptomics preserves this architecture; however, current sequencing-based spatial Single-cell genomics reveal that Alzheimer’s dementia involves the complex interplay of virtually every major brain cell type. Cell-type-specific molecular perturbations modulate signaling A comprehensive atlas of genes, cell types, and their spatial distribution across a whole mammalian brain is fundamental for understanding the function of the brain. Here, using single-nucleus RNA sequencing (snRNA-seq) and Stereo-seq techniques, we generated a mouse brain atlas with spatial information for 308 cell clusters at single-cell resolution, involving over 4
Single-cell RNA-sequencing has already transformed neuroscience. By facilitating detailed comparative and genetic perturbation analyses, it may provide the tools to uncover fundamental mechanisms of neural diversity throughout the tree of life.
This pretreatment is followed by a standard single-cell RNA-seq protocol (D), including reverse transcription, amplification, and sequencing, and generates the Patch-seq sample (lower); the gene expression profile of the Patch-seq sample can be used to map the cell onto the above transcriptomic reference atlas (the red point in E While single-cell RNA abundances in the developing brain have been characterized by single-cell RNA sequencing (scRNA-seq), single-cell protein abundances have not been characterized.
Single-cell RNA sequencing (scRNA-seq) is changing the way scientists study the brain. Instead of averaging gene expression across thousands or millions of Preparation and isolation of single cell, nuclei isolation, RNA isolation, single cell RNA amplification procedures, library construction for sequence analysis and RNA sequencing. In this issue of Nature Neuroscience, Kamath, Abdulraouf et al. 6 address this question by using single-nucleus RNA sequencing, transcriptomics and computational tools to study postmortem samples
Durante et al. report the presence of active neurogenic niches in adult humans using single-cell RNA sequencing of the human olfactory neuroepithelium. Data from the olfactory neuroepithelium We perform single-cell RNA sequencing on naïve and fear-conditioned mice, identify 130 neuronal cell types and validate their spatial distributions. Moreover, we showed several cell types linked to other neurological diseases [i.e., spinocerebellar ataxia (SA), hereditary motor neuropathies (HMN)] and neuromuscular diseases [i.e. hereditary spastic paraplegia (SPG), spinal muscular atrophy (SMA)], including an association between Purkinje cells in brain and SA, an association
- Sisley-Paris Eau Efficace Makeup Remover, 10.1 Oz.
- Sitzen Zwa Hochhäuser Auf Der Kellerstiege Und Stricken Mopeds!
- Sinalco Cola Oder Orangenlimonade 1,25 L Angebot Bei Kaufland
- Since My Birthday , Calculate How Many Days You Have Lived
- Simulateur De Revenus Pour Chirurgien-Dentiste En Libéral
- Situation Confuse Au Niger, Les Dernières Informations
- Single-Mode Dispersion , Chapter 4 Chromatic Dispersion
- Sip Bordeaux 33000 – SIP de Bordeaux Centre Amont, Centre des Finances Publiques
- Sind Autoradio Stecker Genormt?
- Single Sign On Login Fails In 8X8 Work For Desktop
- Simson Habicht M54 Motor, Motorrad Gebraucht Kaufen
- Simulador De Comunicação De Cessação De Contrato Pelo Trabalhador
- Simplex Nostalgie, Fahrräder _ Simplex Hollandrad 28" Nostalgie 1960er Oldtimer Fahrrad
- Singularité, Séparation, Relation
- Sitztrainer Aldi Test Preisvergleich